Team : CHOQUET
Research Projects
Chemical Biology and Protein Engineering (Matthieu Sainlos)
Project Leader(s): Matthieu Sainlos
Contact: Matthieu SAINLOS
Tel. (+33) 5 33 51 47 31
 orcid.org/0000-0001-5465-5641
orcid.org/0000-0001-5465-5641
Matthieu Sainlos is a CNRS research director. He obtained his PhD in bioorganic chemistry with Jean-Marie Lehn. Afterwards, he moved to the lab of Barbara Imperiali (MIT) for a post-doctoral stay. In 2009, he joined the team of Daniel Choquet and participated to the creation of the Interdisciplinary Institute for Neuroscience and launched a new research area on the development of chemical biology tools and methods to monitor and control protein-protein interactions.
SCIENTIFIC CONTEXT
The activity is headed by Matthieu Sainlos (DR2, senior researcher) and currently involves Amine Mehidi (post doctoral fellow), Ivana Trivunovic , Emma Pessoa and Manon Darribere (PhD students), Louise Badruna, and Loris Verron (engineers), Margot Berquez (internship). The general line of research consists in exploiting techniques from chemistry and protein engineering in order to develop original tools and approaches to elucidate biological processes involved in the basis of synaptic transmission.
THE LAB

The lab holds most of the instruments dedicated to organic and peptide synthesis, recombinant proteins expression and purification, biochemistry and biophysics. Other activities such as mass spectrometry, molecular biology, cell culture or fluorescence imaging are conducted using the group/institute shared resources or via core facilities.
RESEARCH OVERVIEW
Research is conducted by using a multidisciplinary approach that combines bioorganic and peptide chemistry, protein engineering, biochemistry, biophysics, molecular biology and cellular imaging to investigate the dynamics of protein complexes in the context of synaptic transmission. We are interested in developing methods and tools that will help us understand and define how synaptic proteins are interacting with one another and how these interactions are regulated in physiological conditions or misregulated in pathological conditions. Our projects are focused on protein-protein interactions (PPIs) and we essentially develop two types of strategies to elucidate their implications in biological processes:
-modulation
-monitoring
Modulation is achieved on endogenous systems by the development of biomolecules that will provide control over specific protein interactions with precise spatial and temporal resolution by the use of light. Methods for the observation of proteins and their complexes are developed with the general goal of taking advantage of chemistry and protein engineering techniques in order to generate tools with improved properties (size, nature of the dyes, valence…) that match the constantly improving imaging techniques. In parallel, we also exploit the tools and methods we develop to further elucidate the molecular architecture of the complexes that we are interested in as well as to identify the key molecular elements involved in their regulation.
PROJECTS
BIOMIMETIC LIGANDS
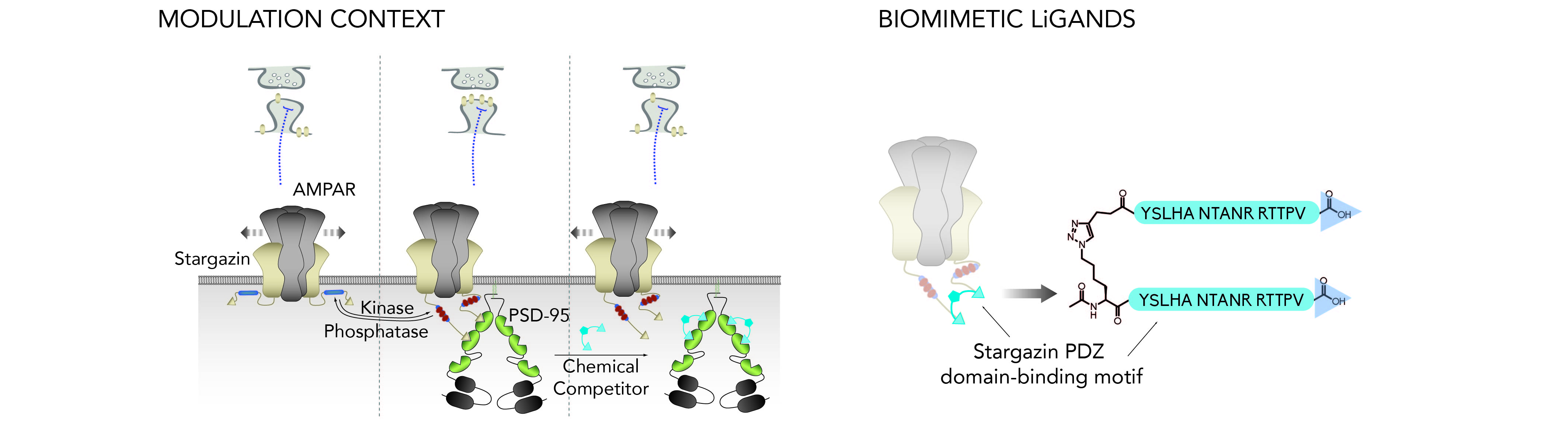
We use biological and structural data to design “biomimetic” peptide-based ligands that incorporate essential structural elements in order to better recapitulate the behavior of the protein complexes of interest (e.g., AMPA receptor complexes, NMDA receptor complexes…) with respect to binding to their partners. For instance, some of our most efficient ligands are composed of multiple PDZ domain-binding motifs that replicate the presence of the multiple subunits within each protein complex. These tools are used to acutely disrupt specific endogenous interactions between the various complexes and their protein partners such as the synaptic PDZ domain-containing scaffold proteins of the PSD-95 family. In parallel, after validation of the tools in cellular settings, we also use these ligands in biophysicial, structural and interactomics studies as models to better define the interactions between the protein complexes they are derived from and their multi-domain-containing partners.
Selected related publications:
Bard et al., Proc. Natl. Acad. Sci. U. S. A. 107, 19561-19566 (2010)
Sainlos et al., Nat. Chem. Biol. 7, 81-91 (2011)
Hass et al., eLife 7, e31755 (2018)
Kellermayer et al., Neuron 100 (1), 106 (2018)
Rosendale et al., Nat. Commun. 10 (1), 4462 (2019)
PHOTOCONTROL OF PPIs 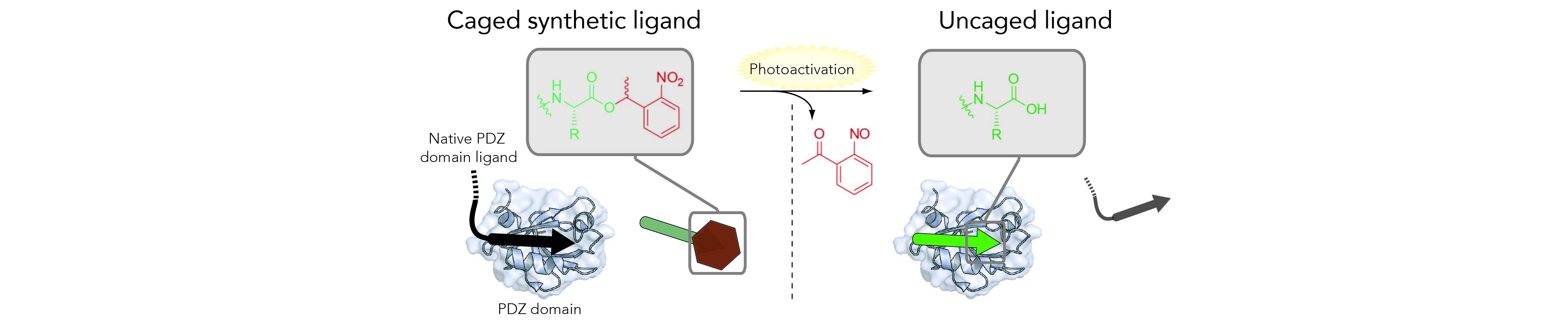
Light provides a powerful approach to achieve high spatial and temporal precision over the effect of a given active molecule when using a photoactivatable or “caged” derivative of that same compound. With the general aim of improving the properties and expanding the range of application of our tools, we develop methods to incorporate and exploit light-sensitive elements (synthetic or protein-based) with the ligands that have been previously validated as efficient PPI modulators or protein binders.
Selected related publications:
Sainlos et al., J. Am. Chem. Soc. 135, 4580-4583 (2013)
DIRECTED EVOLUTION OF PPI MODULATORS
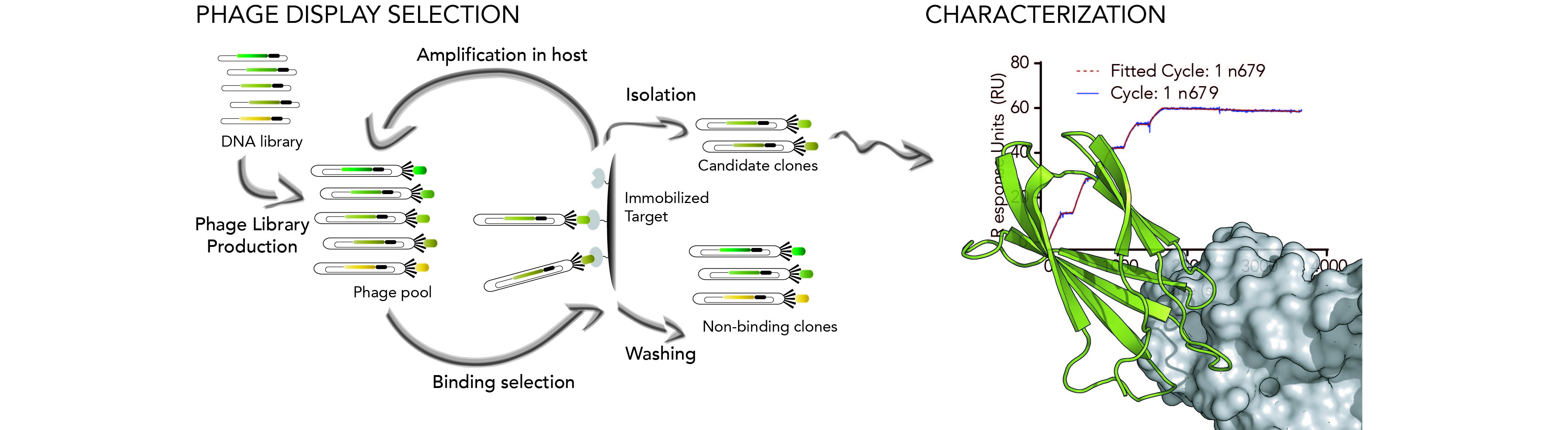
As an alternative to the rational design of peptide-based ligands, we are also developing directed evolution approaches using a phage-based strategy in order to generate small protein domains that are selected for their capacity to efficiently recognize specific domains or PPIs. These tools are ultimately exploited for modulation and imaging purposes.
Selected related publications:
Rimbault et al., Nat. Commun. 10, 4521 (2019)
MOLECULAR TOOLS FOR IMAGING
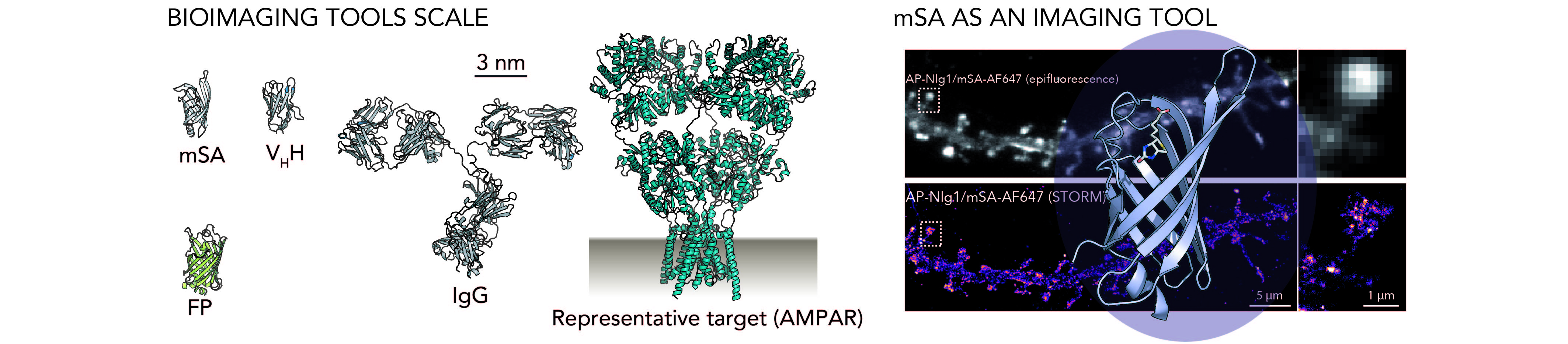
In parallel to the fast improvement of super resolution imaging techniques, there is a growing need for generic protein labeling tools and methods that take advantage of the resulting gain in resolution. In this context, we combine chemical-based approaches and protein engineering in order to develop innovative tools to address the current limitations associated with the existing probes (such as the size, the valence and the nature of the dyes).
Selected related publications:
Chamma et al., Nat. Commun. 7, 10773 (2016)
Chamma et al., Nat. Protoc. 12, 748-763 (2017)
Rimbault et al., eLife, 13:e69620 (2024)
FUNDING
Past and current funding sources include grants from the ANR (CheMoPPI 2013, SynAdh 2014, OptoXL 2016, Stable-FP 2020, AFFLIGEM 2021, Endograal 2023, NEUPOCOP 2023), the Conseil Regional d’Aquitaine (2012) and the BRAIN LabEx.
SELECTED PUBLICATIONS
Engineering paralog-specific PSD-95 recombinant binders as minimally interfering multimodal probes for advanced imaging techniques.
Rimbault, C., Breillat, C., Compans, B., Toulmé, E., Nunes Vicente, F., Fernandez Monreal, M., Mascalchi, P., Genuer, C., Puente-Muñoz, V., Gauthereau, I., Hosy, E., Claverol, S., Giannone, G., Chamma, I., Mackereth, C.D., Poujol, C., Choquet, D. & Sainlos, M.
eLife, 13:e69620 (2024).
Plasmids available from Addgene (link ).
Engineering selective competitors for the discrimination of highly conserved protein-protein interaction modules.
Rimbault, C., Maruthi, K., Breillat, C., Genuer, C., Crespillo, S., Puente-Muñoz, V., Chamma, I., Gauthereau, I., Antoine, S., Thibaut, C., Wong Jun Tai, F., Dartigues, B., Grillo-Bosch, D., Claverol, S., Poujol, C., Choquet, D., Mackereth, C.D. & Sainlos, M.
Nat. Commun. 10, 4521 (2019).
Plasmids available from Addgene (link ).
Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin.
Chamma I., Letellier M., Butler C., Tessier B., Lim K.-H., Gauthereau I., Choquet D., Sibarita J.-B., Park S., Sainlos M. & Thoumine O.
Nat. Commun. 7, 10773 (2016).
Monomeric streptavidin plasmid available from Addgene (#80706 ).
Inhibition of PDZ domain-mediated interactions.
Grillo-Bosch, D., Choquet, D. & Sainlos, M.
Drug Discov. Today Technol. 10, e531-e540 (2013).
Caged Mono- and Divalent Ligands for Light-Assisted Disruption of PDZ Domain-Mediated Interactions.
Sainlos, M., Iskenderian-Epps, W.S., Olivier, N.B., Choquet, D. & Imperiali, B.
J. Am. Chem. Soc. 135, 4580-4583 (2013).
Biomimetic divalent ligands for the acute disruption of synaptic AMPAR stabilization.
Sainlos, M., Tigaret, C., Poujol, C., Olivier, N.B., Bard, L., Breillat, C., Thiolon, K., Choquet, D. & Imperiali, B.
Nat. Chem. Biol. 7, 81-91 (2011).
Full list of publications available here (ORCID)
USEFUL LINKS
- Biochemistry Plateform
- SynDivA (Synthetic gene library Diversity Analysis on Galaxy Europe)
- Proteomic Plateform (MALDI-TOF reservation)
FORMER CHEMICAL BIOLOGY LAB MEMBERS
Dolors Grillo-Bosch, Sara Crespillo, and Virginia Puente (post doctoral fellows), Charlotte Rimbault (graduate student), Isabel Gauthereau, Ségolène Antoine, Camille Genuer, Ellyn Renou, and Cecile Lemoigne (Engineers and technical staff), Ananda Appavoo, Guillaume Dufton, Coraline Thibaut, Marine Dupuis, Laurent Gastambide, Léa Favrelière, Issa Fall, Dephine Cloarec, Esther Gavello Fernandez, Imili Belkacemi, Manon Coursières, Vincent Mallet, Maxence Fontaine, Eva Bloqué, Guillaume Nahmias, Emma Pessoa and Orianne Devarieux (undergrads, LP, M1, M2 and Engineer school).
Fundings
ANR, ERC DynSynMem